Tools and Services
Resources
Communication
The H3ABioNet grant has come to an end after 12 years of funding. Therefore, we will not be updating or adding any new information to the website. However, you can continue to access or find new locations for H3ABioNet generated tools, training, standards and resources on this website. NB: the H3ABioNet helpdesk will remain active and is now accessible as part of the AfriGen-D helpdesk (https://helpdesk.afrigen-d.org./helpdesk/).
Some of the H3ABioNet projects have continued under new funded grants, you can find links to the new project websites below. Where applicable, links have been provided from H3ABioNet to their new locations:
eLwazi Open Data Science Platform (ODSP):
eLwazi ODSP website: https://elwazi.org/
The African Bioinformatics Institute (ABI):
ABI website: https://www.bioinformaticsinstitute.africa/
African Genomics Data Hub (AfriGen-D):
AfriGen-D website: https://afrigen-d.org/


Conference Overview/description:
WiDS Africa is an independent event that is organized by H3ABioNet based WiDS Ambassadors in collaboration with other African based WiDS Ambassadors from MARI and dlab as part of the annual WiDS Worldwide conference organized by Stanford University and an estimated 150+ locations worldwide, which features outstanding women doing outstanding work in the field of data science. All genders are invited to attend all WiDS Worldwide conference events. We are hosting a WiDS Africa regional event to engage our community of diverse women in Data Science. We will watch the WiDS Stanford Livestream as well as feature Africa specific lightning talks, panel discussions, Keynote speech, and end with a networking event.
NB: Registration and Attendance of Conference is FREE
Registrations close 03 March 2021
Intended Audience:
This conference is aimed at; Women in Data Science, Early Career Researchers, Fellows, MSc, PhD trainees, postdocs. Academics, Government and Industry officials. Anyone interested in data science. All genders are welcome.

Conference objectives/outcomes:
• Listen to and engage with a diverse group of women in data science through panels and talks
• Networking
• Advocacy
• Promote Data Science in Africa especially for women
Prerequisites:
None. Just be interested in Data Science and promoting gender diversity in the field.
Dates for the Conference:
Monday 8 March 2021
Conference registration opens:
04/02/2021
Classroom registration closes:
03/03/2021
Notification date:
Immediately after registration
Participant registration:
Registrations are now open, please register here. Registration close 3 March 2021 at 11:59 pm CAT.
Workshop organisers:
African based WIDS Ambassadors — H3ABionet: Paballo Chauke, Nicky Mulder, Caleb Kibet. dlab: Mahadia Tunga. MARI: Jacki O'Neill. BMGF: Amel Ghouila
Workshop Sponsors:
WiDS Stanford University
WIDS
Keywords:
Data Science, Women in Science, International Women's Day
Skill level of training:
Beginner to Advanced
Language:
English
Credential awarded:
N/A
Type of meeting:
Virtual
Conference Programme:
Monday 8 March 2021: African WIDS Regional Programme
|
Time (CAT) |
Session Title |
Speaker/s |
Facilitator/ Moderator |
|
14 :00 – 14 :10 |
Welcome and Introductions |
Nicky Mulder and Mahadia Tunga |
Mahadia Tunga |
|
14 :10 – 14 :30 |
Opening Talk |
Amel Ghouila (BMGF) |
|
|
14 :30– 15 :00 |
Panel Discussion on Impacts of the Pandemic on women in data science |
Speakers:
Chenai Chair (Mozilla)
Rahab Wangari(Hepta Analytics)
Ruthbetha Kateule(UD)
Dominique Anderson (SANBI) |
Verena Ras |
|
15 :00- 15 :05 |
Break/breather |
|
Verena Ras |
|
15 :05-15 :12 |
Graduating during a pandemic |
Samar Elsheikh |
Mahadia Tunga |
|
15 :12- 15 :30 |
Launching your career as a data science consultant 2 speakers 7 mins each (14 + 3min Q&A) |
Hellen Maziku (UD) Catherine Cress |
Mahadia Tunga |
|
15 :30- 16 :00 |
KEYNOTE PRESENTATION |
Mamokgethi Phakeng (UCT VC) |
Mahadia Tunga |
|
16:00- 16 :10 |
Young girls in DS- passing the baton and being black/POC in DS |
Jenea Adams (BWCB)
Winfred Gatua (PU) |
Amel Ghouila |
|
16 :10-16 :15 |
Closing and Thank you remarks |
|
Nicky Mulder |
|
16 :15-17 :00 |
Networking session |
Everyone |
Olawumi, Verena, Amel, Mahadia and Jacki |

Speakers Country of Origin:
Logos of organizers/ collaborators:



H3ABioNet provides access to experts from a variety of domains to help answer any bioinformatics related questions and provide support to various H3Africa and non H3Africa projects that might be struggling with the analysis and planning of their experiments. The H3ABioNet helpdesk system accepts requests from the H3ABioNet web portal registered and non-registered users and can be accessed from here.
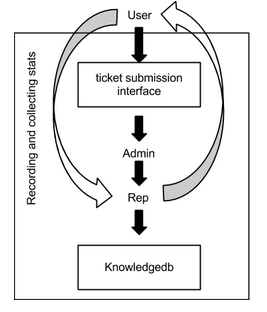
Process flow of a user submitted query to the H3ABioNet helpdesk.
A User submits a H3ABioNet helpdesk query via the submission interface which is assigned to an Admin user who will check what Category the query belongs to, who is available to answer the query and assign the query to that Rep. The Rep will answer the query or request the User to submit more information in terms of providing context of the problem if necessary. The H3ABioNet helpdesk Rep and User will work together until the query has been resolved.
Queries can by users can currently be submitted to the H3ABioNet helpdesk using 3 different methods:
Directly from the H3ABioNet helpdesk web portal. This option is available to H3ABioNet registered and non registered users. The difference between a registered user and a non-registered user is the former can log into the H3ABioNet website and view the history of all the tickets that they have submitted and their status. For the latter, the status of each submitted ticket can only be queried individually via a simple interface.
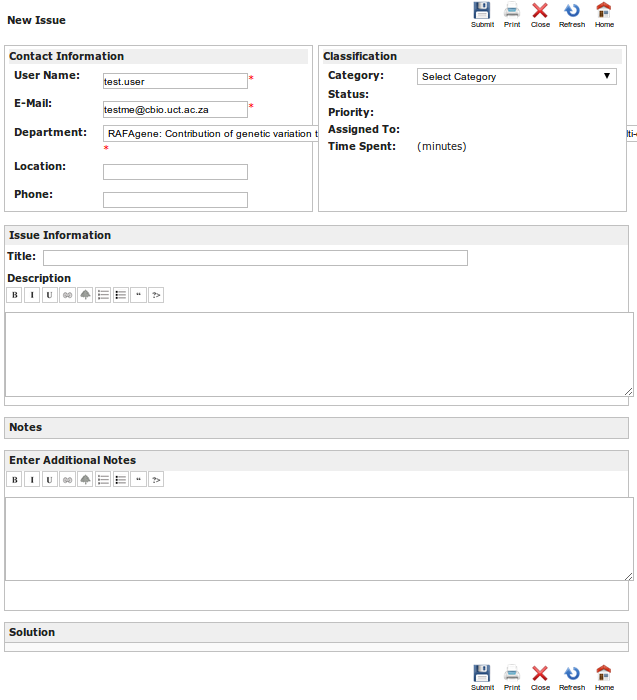
Screen shot of the H3ABioNet helpdesk query submission interface used to submit a question to H3ABioNet by a user.
An H3ABioNet helpdesk Admin can create ticket on behalf of a User upon a request for any assistance and provide the ticket number to the User for subsequent communication
Tickets can automatically be created by sending an email to (although this is not recommended).
The user selects the Category their query falls within, chooses the Department (H3Africa project they are part of or "None" if they are not part of an H3Africa project), enters their email address and provides a title in the form of a question. The user then inputs the details of the query they have into the description box and clicks on "Submit". The user will receive an email acknowledging their submission which will contain an ID number (ticket number used for tracking by the H3ABioNet helpdesk system).
Additional information can be added to a ticket by sending an email directly to and quoting [psn:X] in the subject line where X is equal to the ticket number.
The H3ABioNet helpdesk Admin will receive the query and based on the category entered by the user and will assign the query / ticket to an H3ABioNet helpdesk expert (a Rep). The Rep or Admin will also assign a priority to the submitted ticket which could be:
The Rep will then endeavour to answer the query submitted by the user via the H3ABioNet helpdesk interface. While the H3ABioNet helpdesk Rep is working on the user submitted query, the ticket itself has a status which can be modified by a Rep or Admin. The various options for the status of an H3ABioNet helpdesk ticket are:
These statuses will change during the life cycle of the submitted query until it has been fully answered and the ticket associated to the query can be closed.
There will usually be some communication between the Rep and user on the query that was submitted before a final solution is reached. The H3ABioNet helpdesk system enables the tracking of various threads of communication revolving around that query via the ID / ticket number via a convenient web based interface. An email is also sent to the Rep and the user whenever an update to the submitted query is made such as when a Rep or user replies on a ticket (in the Enter Additional Notes field) so both the user and Rep can attend to the update.
An example of interaction and additional notes taken between a user and Rep.
Once a final solution for the problem has been reached to everyones' satisfaction, the Rep can enter a summary of this solution within the "Solution" box of their interface and close the ticket. The information / final solution provided will be entered into the Knowledge base portion of the H3ABioNet helpdesk system upon the Rep's and Admin's discretion for use in generating FAQs and also providing solutions to any similar queries that might be submitted in the future by another user. The Knowledgebase is a searchable database of user submitted queries to which a solution has been achieved and can only be queried by Reps and Admins.
Currently attachments cannot not be sent to the H3ABioNet helpdesk when a user creates a ticket. A user or admin would first need to create a ticket and then only can a user submit an attachment to the H3ABioNet helpdesk by sending anattachments can be send to and quoting [psn:X] in the subject line.
Genome Detective is a web-based, user-friendly software application to quickly and accurately assemble all known virus genomes from next-generation sequencing datasets. This application allows the identification of phylogenetic clusters and genotypes from assembled genomes in FASTA format. Since its release in 2019, we have produced a number of typing tools for emergent viruses that have caused large outbreaks, such as Zika and Yellow Fever Virus in Brazil. Here, we present The Genome Detective Coronavirus Typing Tool that can accurately identify the novel severe acute respiratory syndrome (SARS) related coronavirus (SARS-CoV-2) sequences isolated in China and around the world. The tool can accept up to 2,000 sequences per submission and the analysis of a new whole-genome sequence will take approximately one minute. The tool has been tested and validated with hundreds of whole genomes from ten coronavirus species, and correctly classified all of the SARS-related coronavirus (SARSr-CoV) and all of the available public data for SARS-CoV-2. The tool also allows tracking of new viral mutations as the outbreak expands globally, which may help to accelerate the development of novel diagnostics, drugs, and vaccines to stop the COVID-19 disease. Read more.
Source: Bioinformatics, 2020.
1. Can I submit a ticket even if I am not part of the H3Africa consortium?
Yes, anyone can submit a query. This service is open to all scientists in Africa on the project name please choose “Other”.
2. Why do I need to fill in all the information requested when I submit a ticket?
We need this data for reporting purposes. It will not be used against you in anyway.
3. Can I ask one of the HelpDesk members to perform an analysis for me?
We would prefer to keep this as an informational HelpDesk. We would suggest that you set up a collaboration agreement between you and the HelpDesk rep you are working should you need some analysis done for you.
4. Can I submit an attachment as part of the ticket?
A user or admin would first need to create a ticket and then only can a user submit an attachment via email. Send attachments to and quote your ticket number in the subject line.
Page 1 of 3
Tools and Services
Communication

