H3ABioNet provides guidelines on the use of clinical data, data management and data standardization
Data & Standards
Phenotype Data Collection Standards
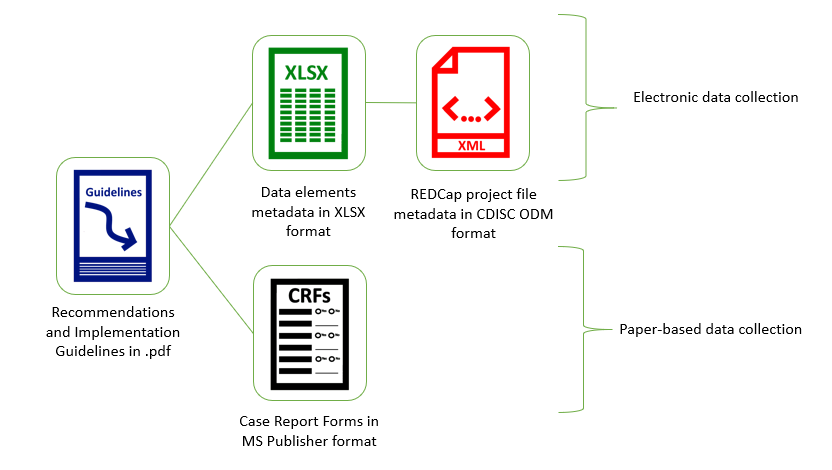
H3ABioNet has built resources for the developed modules in the form of a REDCap Project Template. This offers an XML file which can be uploaded in any data management platform which accepts the format as a complete project incorporating a Data Dictionary with the standardised data definitions and formats for all phenotypes contained in a module. Each module is accompanied by a guidelines document which describes the module and instructions on how to collect data in the module. Finally, selected modules also have an associated .pdf CRF file, which can be used for data collection directly from research participants.The benefits of using these modules are multi-fold:
- The modules are all aligned with existing, global data collection standards.
- The modules are specifically adapted for biomedical phenotype data collection in Africa.
- The modules are highly interoperable, with variables mapped to existing and maintained ontologies.
The aforementioned data collection tools are further described below, and can also be accessed through H3ABioNet’s GitHub or Zivahub. In addition, more info on the history and methodology of work, as well as info on how to acknowledge and cite our work can be found in the documentation. A related publication has also been released.
- Developing Clinical Phenotype Data Collection Standards for Research in Africa. Zass et al., (2024).
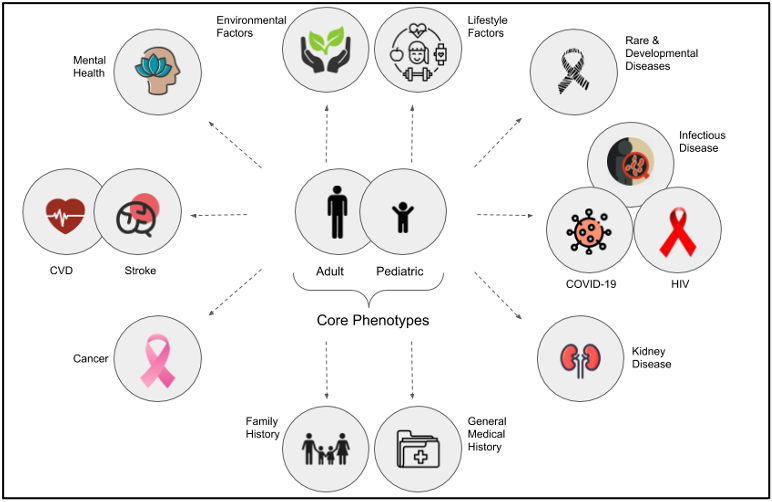
Core Phenotypes - Standard CRF
The Core Phenotypes are a recommended set of phenotype data elements broadly applicable across Africa - dubbed the “Std. CRF”. Data collected with the Std. CRF provides phenotype data complementing the genomic data generated by H3Africa. The core phenotypes are customised for collection in both adult and pediatric research participants.
- Core Phenotypes (Adult): REDCap Project Template, Data Dictionary, Guideline
The Std. CRF pdf files can also be downloaded individually and used for on-site data collection:
OR, select individual pdf forms from the Std. CRF
- Demographics
- Smoking Status
- Alcohol Consumption
- Drug Use
- Anthropometrics
- Blood Pressure
- Urine Test Results
- Kidney Disease
- Prescribed Medications
- CVD
- Stroke History
- Diabetes
- HIV
- Dyslipidemia
- Cancer
- Infectious Diseases
Domain-Specific Modules
A range of extendable domain-specific modules have been developed and can be used in conjunction with the aforementioned Core Phenotypes. These modules can be employed to promote comprehensive and harmonized data collection within specific research domains. Domain-specific modules are available for download via H3ABioNet’s GitHub or Zivahub and include:
- Cancer
- Cardiovascular Disease
- COVID-19
- Environmental Exposures
- Family History
- General Medical History
- Infectious Diseases
- Kidney Disease
- Lifestyle Factors
- Microbiome
- Rare & Developmental Disorders
- Stroke




