One important aspect of managing emerging infections is identifying chains of transmission and assigned cases to clusters of infection. A case in point is South African trade union leader Zwelinzima Vavi, who spent a few days in hospital with the new coronavirus disease. He stated that he “had no idea” where he may have contracted the virus and was scrutinising his travel history for clues.
Peter van Heusden, University of the Western Cape
This experience is hardly unusual, and would be more so for South Africans using public transport and living in crowded circumstances. To ensure that scientists can trace people’s contacts, stronger systems of disease surveillance are needed – ones that draw on genome sequencing.
The first two SARS-CoV-2 virus sequences from the African continent were published by ACEGID in Ede, Nigeria and INRB in Kinshasa, Democratic Republic of the Congo, in early March. These labs have played significant roles in research on and fighting Ebola. With strong research links to the US, they represent both African success stories and outposts of a “health security” regime focused on emerging pathogens “over there” (in Africa) that might threaten the lives “over here” (in the US). The global health security model is based on this flow of disease. However, COVID-19 has reversed the narrative.
The US and Europe, rather than African countries, are among the epicentres of the pandemic. If the global health security model monitors disease trajectories along the lines of networks of exchange, the model has worked largely as designed. Information and techniques have been shared, especially between Western scientists and health authorities, in an unprecedented fashion since the start of 2020.
In South Africa, the National Bioinformatics Institute (SANBI) recently collaborated with the National Institute for Communicable Disease (NICD) to produce the first SARS-CoV-2 viral genome collected in South Africa. Soon after this the Kwazulu-Natal Research Innovation and Sequencing Platform (KRISP) published five further genomes.
The variation captured in these genomes, when compared to genomes sampled elsewhere, provides a fingerprint that might be associated with a particular virus – and so, a patient with a particular cluster of transmission.
The first use of this kind of finger printing, more formally known as genomic epidemiology, was to trace the source of the anthrax used in the 2001 anthrax letter attacks in the US. In South Africa similar techniques were used to identify the source of a 2017-2018 listeriosis outbreak.
Worldwide, genomic surveillance techniques are proving useful in tracking the spread of COVID-19 and South Africa is well positioned to adopt them within its public health system response.
How it works
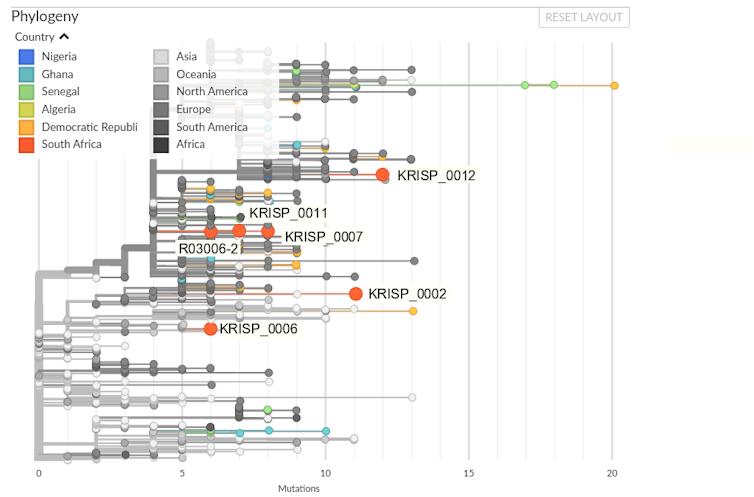
All six SARS-CoV-2 genomes produced in South Africa thus far have been submitted to the GISAID data repository and from there incorporated in the Nextstrain platform. Nextstrain uses both sequence data and sample metadata to produce a phylogenetic tree and map of the virus’ global spread. Figure 1 shows the phylogenetic tree with African and, in particular South African, sequences highlighted.
In reading the phylogenetic tree remember that the SARS-CoV-2 virus has a genetic code of about 29000 letters of RNA. This RNA is essential to describing the virus’ proteins and is replicated millions of times as the virus spreads within a person and transmits to a new host. While SARS-CoV-2 is assisted in its replication by a “proofreading” exonuclease, mutations accumulate in the virus at a rate estimated at 0.8x103 per nucleotide site per year.
These mutations can involve the addition, removal or rearrangement of parts of the genome sequence. But single nucleotide variations – the change, for example of a guanine (G) to a cytosine (C), are the most informative for current phylogenetic techniques.
The evolution represented by the Nextstrain phylogeny is all considered with regards to the “reference” sequence of SARS-CoV-2, derived from a sequence collected on 26 December 2019 in Wuhan, China. The South African R03006-2 sequence differs from the reference sequence at six positions that bear some similarity to virus sequences collected in Europe.
While this is a relatively modest signal and must not be over-interpreted, it is clear from the phylogenetic tree that the South African virus sequences are found in four different places with regards to the global distribution of virus. This indicates multiple imports of COVID-19 into the country.
The situation with R03006-2, KRISP_011 and KRISP_007 is more difficult to discern: are they part of a transmission chain? Figure 2 presents a closer look.
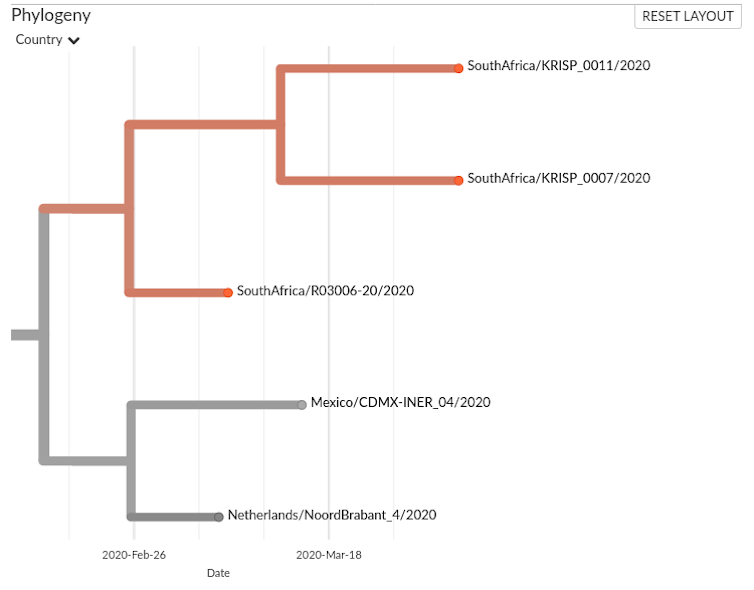
The R03006 sample was collected on 7 March, and the KRISP samples three weeks later, on 1 April. The sequences are very similar and there could be a link, but the small number of sequenced samples (six out of 1380 cases reported in South Africa by 1 April, and 8000 sequenced out of nearly 2 million cases worldwide) hampers our ability to draw conclusions. Perhaps R03006 and the KRISP samples illustrate transmission from another person whose virus is not present in the data.
Genomic epidemiology can aid in understanding disease transmission, but it is not enough on its own. It needs to be combined with field epidemiological data for correct interpretation. The more sequences we have to work with, the more bioinformaticians can assist their colleagues in understanding transmission and the structure of the COVID-19 disease outbreak.
What’s missing
Surveying SARS-CoV-2 sequencing on the African continent reveals a weakness. Of the first 87 virus genomes published, 86 emerged from research institutes. While many of these have strong links with national health systems, they also reflect both the weakness of African governments’ investment in science and a disconnect between genomic science and public health. Silos also exist between national initiatives, while the continent faces a singular disease threat.
In this context, the launch of a Pathogen Genomics Intelligence Institute (PGII) by the Africa CDC in 2019 was prescient.
Surveillance is part of the ‘hardware’ of health systems’ preparedness for emerging infectious diseases. The South African response to COVID-19 clearly aligns with this vision, but it is crucial that virus sequencing and genomic epidemiology be deployed within the public health response to understand and counter disease transmission in the coming months.![]()
Peter van Heusden, Researcher, University of the Western Cape
This article is republished from The Conversation under a Creative Commons license. Read the original article.




